The Cupid Peptide Company
EXPERTS IN CELL PENETRATING PEPTIDE
CUSTOM DESIGN AND MANUFACTURE
P53
P53 is a tumour suppressor protein that regulates the expression of a wide variety of genes involved in Apoptosis, Growth arrest, Inhibition of cell cycle progression, Differentiation and accelerated DNA repair or Senescence in response to Genotoxic or Cellular Stress.
As a transcription factor, p53 is composed of an N-terminal Transactivation Domain, a central specific DNA Binding Domain, and a C-terminal Tetramerization Domain, followed by a Regulatory Domain rich in basic Amino acids. Having a short half-life, p53 is normally maintained at low levels in unstressed mammalian cells by continuous ubiquitylation and subsequent degradation by the 26S Proteasome. Non-phosphorylated p53 is ubiquitylated by the MDM2 (Mouse Double Minute-2) ubiquitin ligase. MDM2 binding inactivates p53 by at least two mechanisms. First, MDM2 binds to the transactivation domain of p53, precluding interaction with the transcriptional machinery. Second, this binding mediates the covalent attachment of ubiquitin to p53. Ubiquitylated p53 is then degraded by the Proteosome. Thus MDM2 acts as a major regulator of the tumor suppressor p53 by targeting its destruction. When the cell is confronted with stress through DNA damage, Hypoxia, Cytokines, Metabolic changes, Viral infection or Oncogenes, however, p53 ubiquitylation is suppressed and p53 accumulates in the nucleus, where it is activated and stabilized by undergoing multiple covalent modifications including Phosphorylation and Acetylation (1-4).
Reviews
(1) Another fork in the road--life or death decisions by the tumour suppressor p53. Carvajal LA, Manfredi JJ. EMBO Rep. 2013 May;14(5):414-21. doi: 10.1038/embor.2013.25. Epub 2013 Apr 16. Review.
(2) Senescence regulation by the p53 protein family. Qian Y, Chen X. Methods Mol Biol. 2013;965:37-61. doi: 10.1007/978-1-62703-239-1_3. Review.
(3) p53 in health and disease. Vousden KH, Lane DP. Nat Rev Mol Cell Biol. 2007 Apr;8(4):275-83. Review.
(4) Neural precursor cells possess multiple p53-dependent apoptotic pathways. Akhtar RS, Geng Y, Klocke BJ, Roth KA. Cell Death Differ. 2006 Oct;13(10):1727-39. Epub 2006 Mar 3.
P53 interactions
Wikipedia lists over 100 proteins that interact with p53 (1). Of these the most studied is the Mdm2 - p53 interaction between the N-terminal transactivation domain of p53 and the N-terminal region of MDM2. This interaction causes p53 repression which, through differential target gene expression, determines cell fate is of major interest to Cancer therapy.
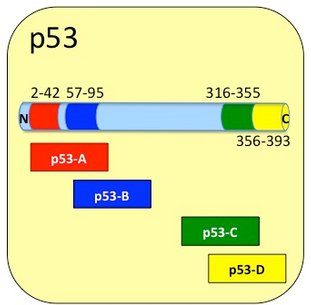
As well as being the site for MDM2 and replication protein A (RPA) interaction, the N-terminus of p53 has many other interesting features. It contains two complementary transcriptional activation domains (transactivation domains or TADs), one at residues 1–42 and another at residues 55–75. p53 transcription is mediated through TAD interaction with a host of general coactivators e.g. TAF9, CBP/p300, GCN5 and PC4. Interplay of such interactions allow p53 to transcribe and regulate a gamut of genes, many specifically related to Apoptosis. The first TAD region is contained in Product: Cupid-p53-A.
Adjacent to the N-terminal region, from residues 64-92, there are five Proline-Rich motifs (-PXXP- motifs) believed to interact with proteins containing SH3 domains (2). This region is contained in Product: Cupid-p53-B.
The central DNA-binding core domain (DBD, residues 102-292) of p53 binds DNA and within this are sites for one zinc atom and the p53 co-repressor LMO3 protein (3).
Towards the C-terminus, at residues 307-355, p53 has an oligomerisation domain which is capable of making a p53 tetramer. Tetramerization is essential for the activity of p53 in vivo. This domain also features a nuclear localization signal at residues 316-325. This region is contained in Product: Cupid-p53-C.
Finally the C-terminal residues, 356-393, which are basic in character, are involved in down-regulation of DNA binding of the central domain and modulation of transcription activity (4). This region is contained in Product: Cupid-p53-D.
References
(1) http://en.wikipedia.org/wiki/P53
(2) The requirement for the p53 proline-rich functional domain for mediation of apoptosis is correlated with specific PIG3 gene transactivation and with transcriptional repression. Venot C, Maratrat M, Dureuil C, Conseiller E, Bracco L, Debussche L. EMBO J. 1998 Aug 17;17(16):4668-79.
(3) LMO3 interacts with p53 and inhibits its transcriptional activity. Larsen S, Yokochi T, Isogai E, Nakamura Y, Ozaki T, Nakagawara A. Biochem Biophys Res Commun. 2010 Feb 12;392(3):252-7. doi: 10.1016/j.bbrc.2009.12.010. Epub 2009 Dec 6.
(4) The C terminus of p53 family proteins is a cell fate determinant. Harms KL, Chen X. Mol Cell Biol. 2005 Mar;25(5):2014-30.
Cupid p53 Cargoes
Human p53, cellular tumor antigen
p53 isoform a
http://www.ncbi.nlm.nih.gov/protein/NP_000537.3
393 amino acids
Cupid Cargo p53-B
DPGPDEAPRMPEAARVAPAPAAPTPAAPAPAPSLSS
Amino Amino acids 57 to 95 (39 amino acids)
Proline-Rich Domain.
Note: Cupid p53 B peptide features the P to R change from the published sequence. This is a common human polymorphism that has been linked to cancer susceptibility.
Cupid Cargo p53-C
PQPKKKPLDGEYFTLQIRFEMFRELNELNEALEIAQA
Amino acids 316 to 355 (40 amino acids)
Oligomerization Domain